
Number: Tu1908
IMPACT OF DIET ON GUT BARRIER FUNCTION AND INFECTION DEVELOPMENT IN A C57BL/6J MOUSE MODEL OF COLITIS
Society: AGA
Track: Microbiome in Gastrointestinal and Liver Diseases
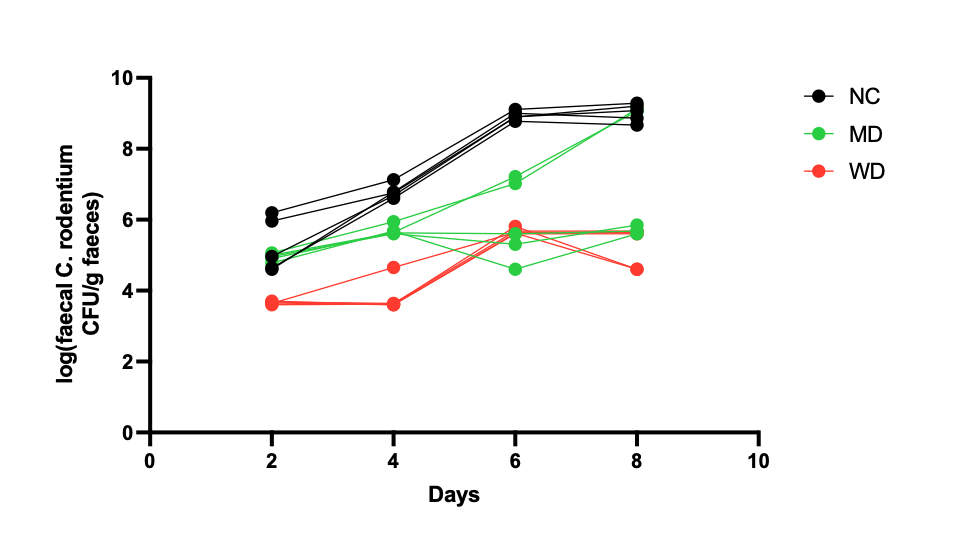
Background: Diet represents a key modulator of the gut microbial composition and metabolic activity, which is believed to influence gastrointestinal (GI) disease initiation and development. Inflammation of the GI tract, often associated to conditions such as inflammatory bowel disease, is linked to adverse health complications and imbalanced microbiota composition. Worldwide, Mediterranean diet (MD) and Western diet (WD) are well known models of healthy and unhealthy eating patterns, respectively, which have also been shown to modulate both GI inflammation risk and the microbiota composition. There is a need to better understand the interplay between gut bacteria and diet in GI disease initiation, in particular the role of bacterial metabolites and host-bacterial co-metabolites. Methods: Wild-type C57BL/6J mice were fed either a normal chow (NC) diet or formulated MD and WD for 2 weeks before inoculation with Citrobacter rodentium(CR) to induce colitis. PBS was used for control animals. Biofluids (urine, faeces, plasma) were collected pre- and post-dietary intervention, as well as daily following inoculation. Gut content and gut tissue were collected post culling. 1H Nuclear Magnetic Resonance (NMR) was used to assess metabolic profiles of biofluids and gut content. Multivariate statistical analysis, namely principal component analysis (PCA) and orthogonal projection to latent structures - discriminant analysis (OPLS-DA), were applied to compare global metabolic profiles. Colony forming units were counted in faeces to assess bacterial growth. Histology markers (crypt length, claudin and goblet cell levels) were evaluated on colon tissue. Results: Dietary interventions resulted in significantly different urinary, faecal, ileal, caecal and colonic metabolic profiles. Dietary interventions resulted in different establishment and growth of CR, with NC showing the highest bacteria counts, followed by MD and WD. Urinary hippurate levels, along with faecal propionate and acetate, were potentially associated with higher bacterial counts. Claudin-2 levels were significantly higher in CR infected WD mice when compared to other groups, whilst claudin-3 and -4 were higher in MD group. Crypt length measurements were significantly higher for CR infected NC mice when compared to other groups. Discussion and Conclusion: Diet can significantly alter the gut metabolic and inflammatory environment leading to differential growth of pathogenic CR in mouse colon. Further examination of histopathology, transcriptional changes to gut tissue and characterisation of immune markers linked to diet and CR pathology is warranted to better understand the relationship between diet, gut microbiota and inflammation in our model. Cytokine profiling of plasma samples, as well as data analysis of bulk RNA sequencing from ileal, jejunal and colonic gut tissue, is currently in progress.
Number: Tu1908
IMPACT OF DIET ON GUT BARRIER FUNCTION AND INFECTION DEVELOPMENT IN A C57BL/6J MOUSE MODEL OF COLITIS
Society: AGA
Track: Microbiome in Gastrointestinal and Liver Diseases
Background: Diet represents a key modulator of the gut microbial composition and metabolic activity, which is believed to influence gastrointestinal (GI) disease initiation and development. Inflammation of the GI tract, often associated to conditions such as inflammatory bowel disease, is linked to adverse health complications and imbalanced microbiota composition. Worldwide, Mediterranean diet (MD) and Western diet (WD) are well known models of healthy and unhealthy eating patterns, respectively, which have also been shown to modulate both GI inflammation risk and the microbiota composition. There is a need to better understand the interplay between gut bacteria and diet in GI disease initiation, in particular the role of bacterial metabolites and host-bacterial co-metabolites. Methods: Wild-type C57BL/6J mice were fed either a normal chow (NC) diet or formulated MD and WD for 2 weeks before inoculation with Citrobacter rodentium(CR) to induce colitis. PBS was used for control animals. Biofluids (urine, faeces, plasma) were collected pre- and post-dietary intervention, as well as daily following inoculation. Gut content and gut tissue were collected post culling. 1H Nuclear Magnetic Resonance (NMR) was used to assess metabolic profiles of biofluids and gut content. Multivariate statistical analysis, namely principal component analysis (PCA) and orthogonal projection to latent structures - discriminant analysis (OPLS-DA), were applied to compare global metabolic profiles. Colony forming units were counted in faeces to assess bacterial growth. Histology markers (crypt length, claudin and goblet cell levels) were evaluated on colon tissue. Results: Dietary interventions resulted in significantly different urinary, faecal, ileal, caecal and colonic metabolic profiles. Dietary interventions resulted in different establishment and growth of CR, with NC showing the highest bacteria counts, followed by MD and WD. Urinary hippurate levels, along with faecal propionate and acetate, were potentially associated with higher bacterial counts. Claudin-2 levels were significantly higher in CR infected WD mice when compared to other groups, whilst claudin-3 and -4 were higher in MD group. Crypt length measurements were significantly higher for CR infected NC mice when compared to other groups. Discussion and Conclusion: Diet can significantly alter the gut metabolic and inflammatory environment leading to differential growth of pathogenic CR in mouse colon. Further examination of histopathology, transcriptional changes to gut tissue and characterisation of immune markers linked to diet and CR pathology is warranted to better understand the relationship between diet, gut microbiota and inflammation in our model. Cytokine profiling of plasma samples, as well as data analysis of bulk RNA sequencing from ileal, jejunal and colonic gut tissue, is currently in progress.



